Custom priors¶
The prior probability is a critical element of Bayes theorem. However, to keep uravu straightforward to use, by default, a broad uniform prior probability is assigned to the Relationship object, or if bounds are present these are used as the limits.
Of course this may be ignored and custom priors may be used (and sometimes it may be necessary that this is done). This tutorial will show how custom priors may be used with uravu.
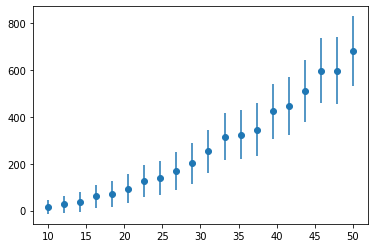
Let’s start, as always, by producing some synthetic data
[1]:
import numpy as np
import matplotlib.pyplot as plt
[2]:
np.random.seed(2)
[3]:
x = np.linspace(10, 50, 20)
y = .3 * x ** 2 - 1.4 * x + .2
y += y * np.random.randn(20) * 0.05
dy = 3 * x
[4]:
plt.errorbar(x, y, dy, marker='o', ls='')
plt.show()
The model for this data is a second order polynomial, below is a function that defines this. The Relationship object is also created.
[5]:
def two_degree(x, a, b, c):
return c * x ** 2 + b * x + a
[6]:
from uravu.relationship import Relationship
[7]:
modeller = Relationship(two_degree, x, y, ordinate_error=dy)
modeller.max_likelihood('mini')
The max likelihood (which makes no consideration of the prior) is found,
[9]:
print(modeller.variable_modes)
[79.94767915 -8.90884397 0.46614102]
The default prior probabilities for these variables with uravu are uniform in the range 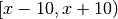 , where
, where 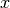 is the current value of the variable.
is the current value of the variable.
However, if you wanted the prior probability to be a normal distribution, centred on the current value of the varible with a width of 1, it would be necessary to create a custom prior function. This function is shown below.
[10]:
from scipy.stats import norm
def custom_prior():
priors = []
for var in modeller.variable_medians:
priors.append(norm(loc=var, scale=1))
return priors
Note that the function returns a list of ‘frozen’ scipy RV objects that describe the shape of the priors.
To make use of these priors, they must be passed to the mcmc or nested_sampling functions as the prior_function keyword argument.
[11]:
modeller.mcmc(prior_function=custom_prior)
100%|██████████| 1000/1000 [01:12<00:00, 13.75it/s]
[12]:
modeller.nested_sampling(prior_function=custom_prior)
2280it [00:34, 65.70it/s, +500 | bound: 2 | nc: 1 | ncall: 18751 | eff(%): 14.826 | loglstar: -inf < -110.472 < inf | logz: -114.157 +/- 0.094 | dlogz: 0.001 > 0.509]
[13]:
print(modeller.ln_evidence)
-114.16+/-0.09
Any scipy statistical function that has a logpdf class method may be used in the definition of priors.